
Background
I am currently a Postdoctoral Researcher at the Cancer Research UK Scotland Institute (formerly known as the Beatson Institute for Cancer Research) in Glasgow, Scotland. My research at the CRUK SI focuses on the use of Salmonella Typhimurium as a bacterial cancer therapy (BCT). Leveraging my previous experience in molecular microbiology and bioinformatics, I am using innovative techniques to investigate the transcriptome of tumour organoids and Salmonella Typhimurium during BCT. As well as tumour organoids, we are currently using mouse models of colorectal cancer (CRC) to understand the host response to CRC and BCT. I am also interested in the role that the gut microbiome plays in colorectal cancer and its role in modulating BCT. My training is as bacteriologist with my previous work focusing on the use of different omics techniques to investigate antibiotic resistance in the gut microbiome with a particular focus on the opportunistic pathogen Enterococcus faecium. My current position allows me to use the skills and knowledge that I have previously acquired in the gut microbiome field to develop novel and innovative cancer therapies.
r.mcinnes@crukscotlandinstitute.ac.uk
Cancer Research UK Scotland (Beatson) Institute
Switchback Rd
Bearsden
Glasgow
UK
Previous positions
- Postdoctoral Research Fellow, University of Birmingham, England, 2022-2024.
Qualifications
- PhD Antimicrobials and Antimicrobial Resistance, University of Birmingham, England, 2022.
- MRes Antimicrobials and Antimicrobial Resistance, University of Nottingham and University of Birmingham, England, 2018.
- BSc Biochemistry and Microbiology Hons (First Class), University of Strathclyde, Scotland, 2017.
Research Interests
Host-bacterial interaction
Bacterial cancer therapy
Bacterial Cancer Therapy (BCT) is the earliest form of immunotherapy that uses bacteria to directly and indirectly kill cancer cells. A plethora of different bacteria have shown efficacy in treating cancer, however the most widely used organism is Salmonella Typhimurium (STm). It’s ability to grow in both aerobic and anaerobic environments, ease of genetic manipulation, capacity for intracellular invasion, and intrinsic tumour homing ability make it an excellent therapeutic candidate. Auxotrophic non-virulent strains and strains engineered to carry anti-cancer payloads have been developed which have shown efficacy in different models. In vitro and in vivo models have been developed that can be used to test the efficacy of BCT. These include organoids derived from tumours of the colon and small intestine that provide a simple and high-throughput method to study the impact of STm on tumour cells. However, organoids lack the critical immune aspect needed to fully elucidate the mechanism of BCT. Mouse models of colorectal cancer including AOM-DSS and ApcMin/+ are helping to unravel the complex interplay between BCT, cancer cells, and the immune compartment.
My current interests in Bacterial Cancer Therapy include:
- The competition between STm and immune cells for tumour metabolites.
- The transcriptional response of STm and tumour cells during BCT.
- The genetic manipulation of STm to increase its efficacy against cancer cells.
- The interaction of the gut microbiome with colorectal cancer cells.
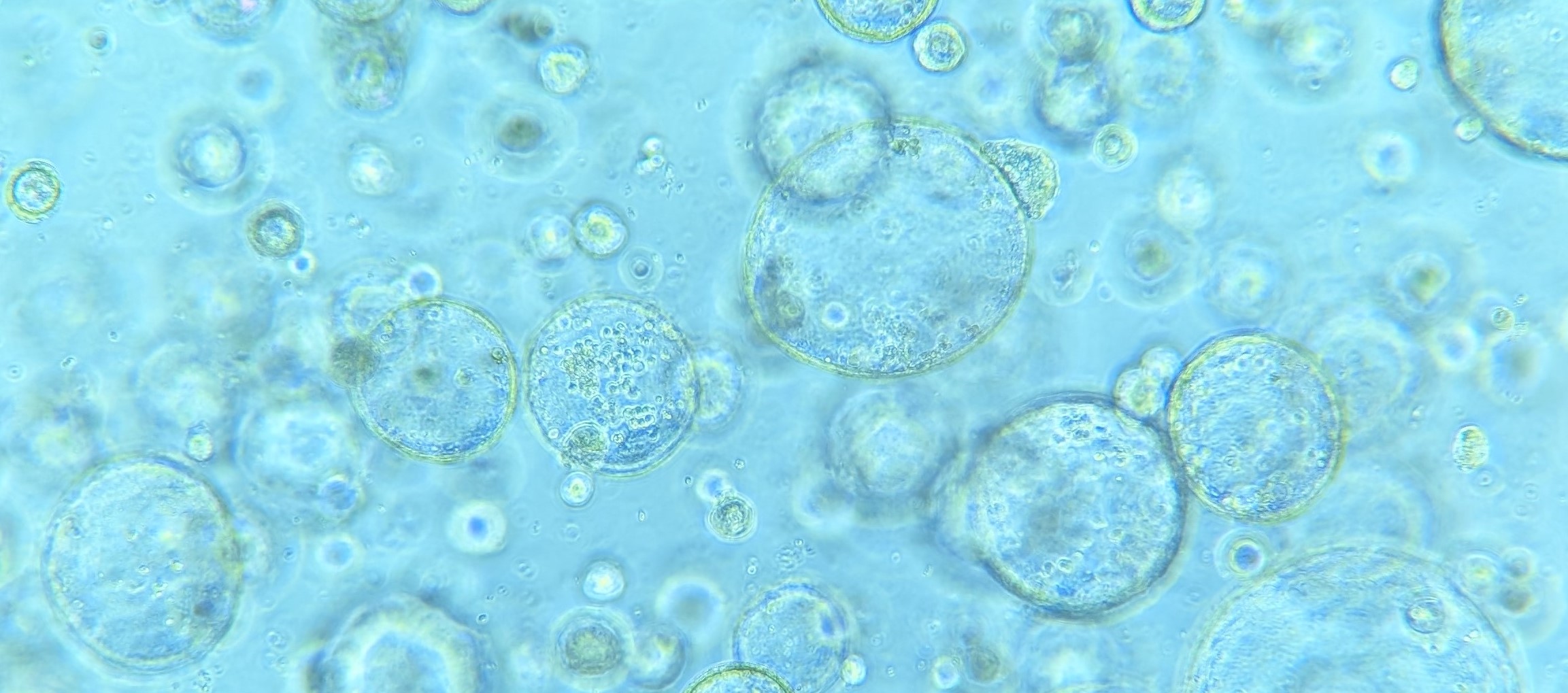
Colon tumour organoids grown within a Matrigel dome.
Antibiotic resistance
Enterococcus faecium
Enterococcus faecium is a low-GC Gram-positive opportunistic pathogen that is associated with the hospital environment. This organism is a major cause of urinary tract infections, endocarditis, and bloodstream infections in humans. It is a particularly hardy organism that can survive in nutrient limited conditions and is resistant to many clinically important antibiotics. E. faecium strains are increasingly resistant to the glycopeptide antibiotic vancomycin. Resistance to vancomycin is conferred by a number of well characterised vancomycin resistance genes of which vanA and vanB are the most common. The vancomycin resistance genes are found on mobile genetic elements including conjugative plasmids and integrative and conjugative elements which means that they can be readily transferred between strains.
My current interests in Enterococcus faecium include:
- Vancomycin resistant E. faecium (VRE) hospital outbreaks.
- Alternative mechanisms of vancomycin tolerance.
- Vancomycin-variable E. faecium strains.
- Simultaneous carriage of different sequence types.
- Horizontal transfer of vancomycin resistance genes.

SEM image of Enterococcus faecium.
Human gut microbiota
The gut microbiota is the community of microorganisms (bacteria, fungi, viruses etc) that live within the human gastrointestinal tract. It has many important functions including colonisation resistance, nutrient metabolism, and immune stimulation. The gut microbiota contains opportunistic pathogens that are normally kept at low levels by the other gut commensals. However, in cases of immunodeficiency or perturbation by antibiotics, these opportunistic pathogens can “bloom” and become the major component of the gut. The increased abundance of these organisms makes it more likely that they can cross into the bloodstream and cause disease. It is also hypothesised that the commensal bacteria within the gut microbiota can act as a reservoir for antibiotic resistance gene which can be transferred to the opportunistic pathogens.
My current interests in the human gut microbiota include:
- Horizontal gene transfer of antibiotic resistance genes within the gut.
- Linking antibiotic resistance genes to their bacterial hosts.
- The contribution of the gut microbiota to human disease.
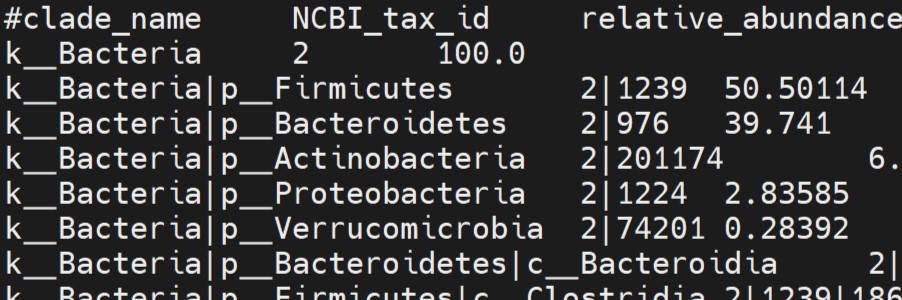
A taxonomic output from MetaPhlAn3.
Expertise
Selected Publications
McInnes, R.S., Snaith, A.E., Dunn, S.J., Papangeli, M., Hardy, K.J., Hussain, A., Van Schaik, W. (2024) ‘Integration of vanHAX downstream of a ribosomal RNA operon restores vancomycin resistance in a susceptible Enterococcus faecium strain’, npj Antimicrob Resist, 2, 2; doi:10.1038/s44259-023-00017-0.
McInnes, R.S., Uz-Zaman, M.H., Alam, I.T., Ho, S.F.S., Moran, R.A., Clemens, J.D., Islam, M.S., Van Schaik, W. (2021) ‘Metagenome-Wide Analysis of Rural and Urban Surface Waters and Sediments in Bangladesh Identifies Human Waste as a Driver of Antibiotic Resistance’, mSystems, 6(4), pp. e00137-21. doi:10.1128/mSystems.00137-21.
McInnes, R.S., McCallum, G.E., Lamberte, L.E., Van Schaik, W. (2020) ‘Horizontal transfer of antibiotic resistance genes in the human gut microbiome’, Current Opinion in Microbiology, 53, pp. 35–43. doi:10.1016/j.mib.2020.02.002.
Extracurriculars
Gardening
I have been a keen gardener all of my life, but like many others it became a real passion (and escape) during the COVID-19 lockdowns. During COVID I was lucky enough to live in a flat with a large balcony, which gave me the opportunity to grow a wide variety of fruit, vegetables, and flowers. When I moved back to Scotland, I bought a house with a small front and back garden that I'm currently transforming into a cottage style garden. I was also able to install a small greenhouse that will allow me to grow some more exotic fruit and vegetables than the Scottish climate would normally allow. If you are interested in my gardening exploits you can see what I have been (and will be) upto on the Instagram page The Wee Garden Grower


